This post is a quick announcement [fixed spelling] of updates to 3 projects and the creation of an umbrella organization to house them: ferritin-bio
Ferritin
Rust library for handling proteins.
- Tagged a v.0.1
- Candle-native AMPLIFY model
- ONNX versions of LigandMPNN encoder, LigandMPNN decoder, and ESM2.
- Conversion of PDB files to matrices that can be used for any of the three models [fixed spelling] above.
Protein-Language-Models
Collation of model info.
Fennec
Local Application for running protein language models on one’s [added apostrophe] own hardware. Kick the tires here!
Homepage
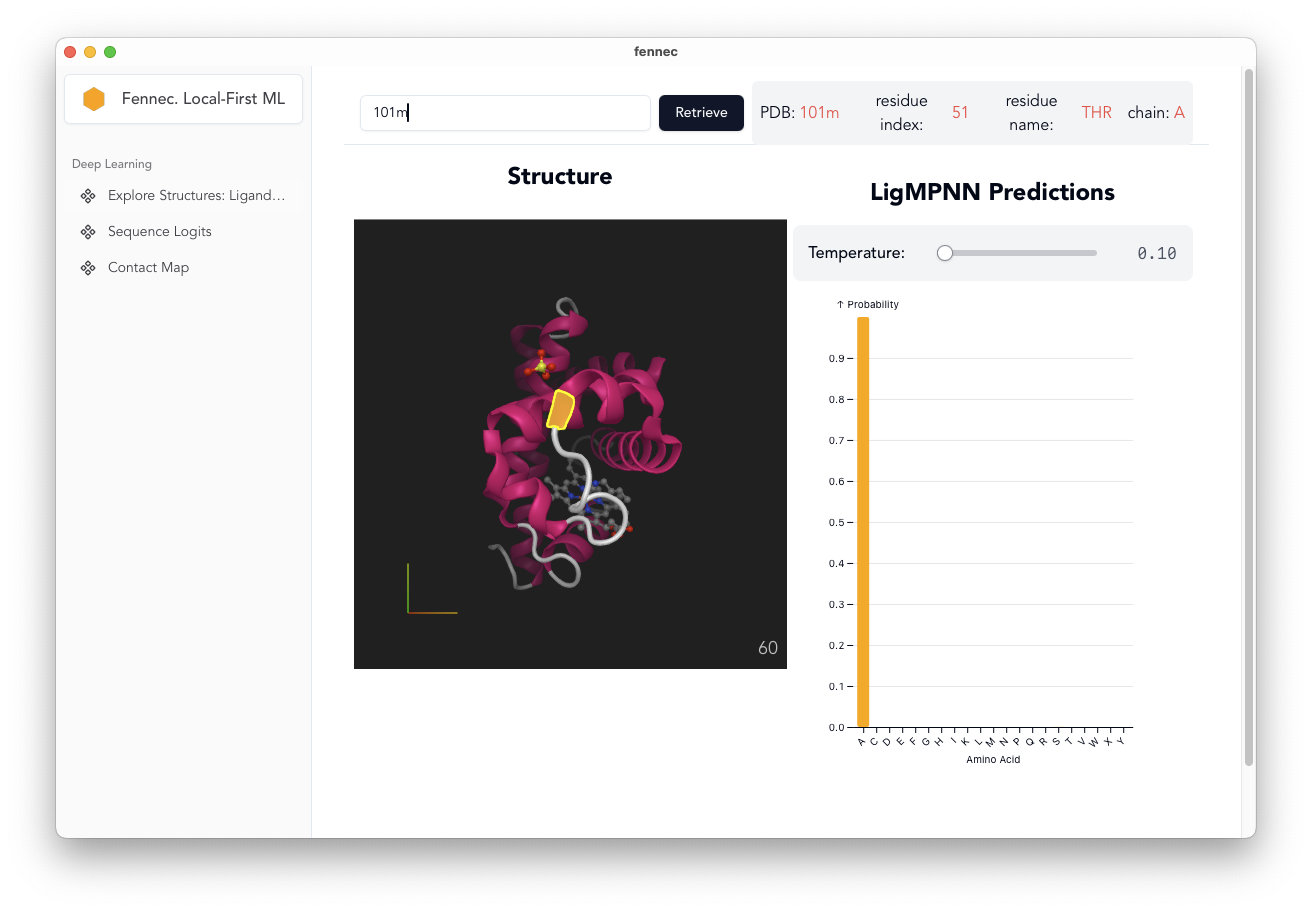
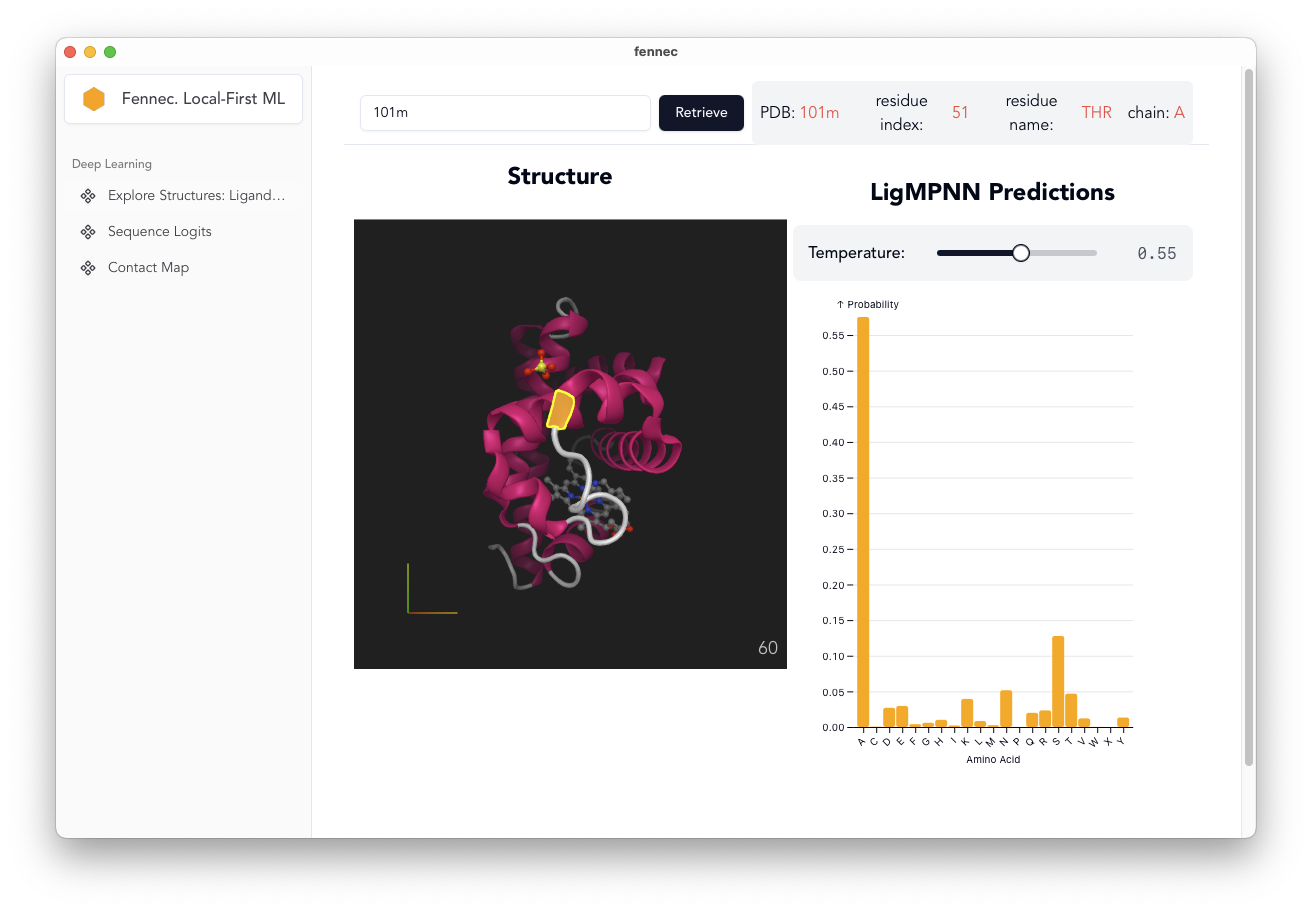
Residue Specific LigandMPNN Predictions
- LigandMPNN weights converted to ONNX and stored on HFHub
- Click Protein:
- encode the whole protein
- decode the predictions per-residue.
- can optionally adjust [fixed spelling] decode temperature
- only works for single chain proteins at the moment

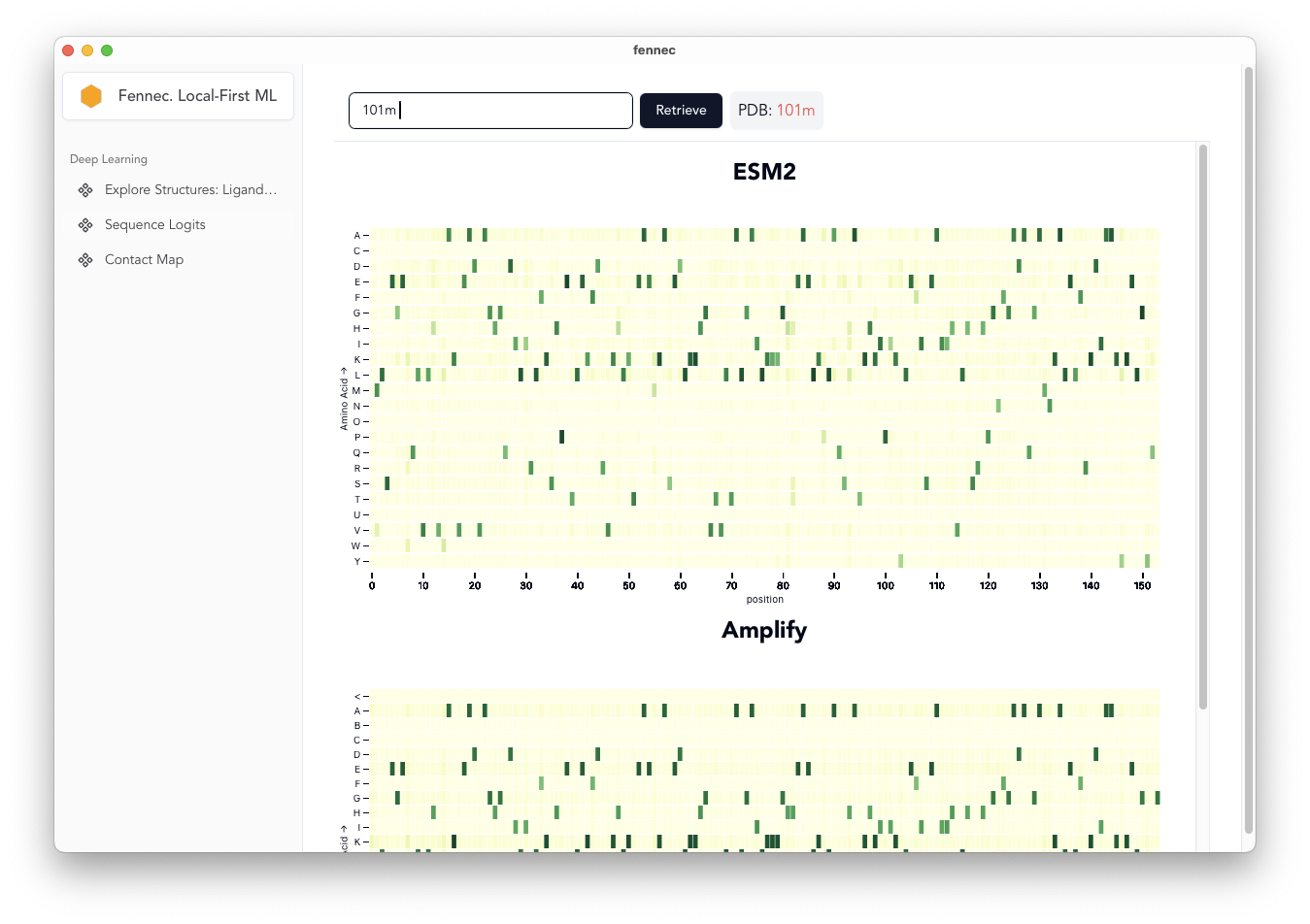
All Sequence Prediction
Return predicted probabilities [fixed spelling] of sequences using masked sequence models ESM2 and AMPLIFY.
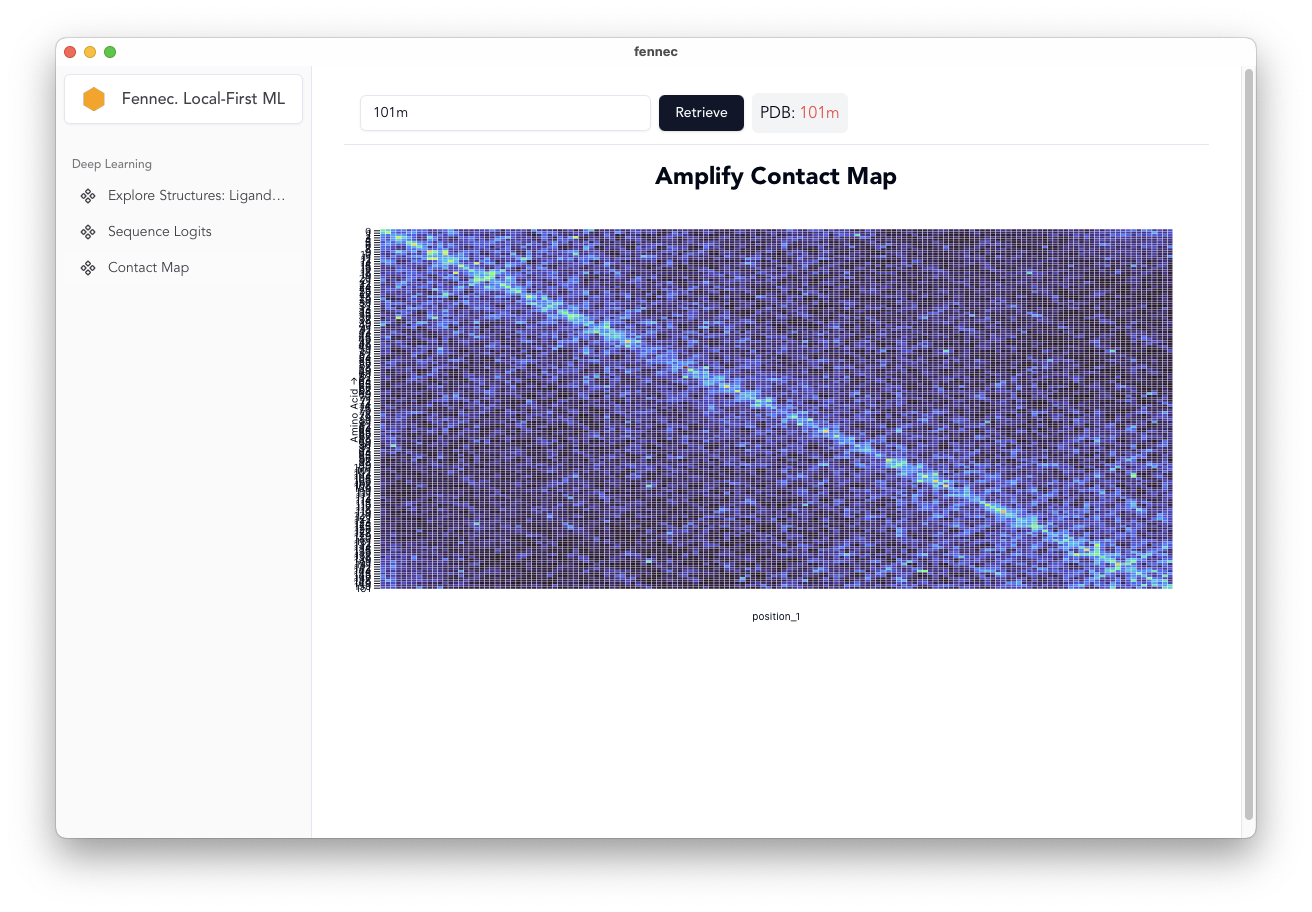
Contact Map Prediction [fixed spelling]
- Use AMPLIFY and return the attention heads.
- calculate the pseudo-contact map/